Gilbert and Stephens Microbiology of the Built Environment Nature Reviews Microbiology 2018
Introduction
Equally over 90% of the average 24-hour interval is spent indoors, in our homes, workplaces and/or travel, indoor exposure to dust and its intrinsic biological, physical and chemic entities represents one of modern society's greatest exposures to potentially harmful substances. A meaning component of house dust is of biological origin, including microbes and their secretions, dead skin cells, dander, hair and respiratory secretions (Morawska and Salthammer, 2003). The ingress of outdoor particulate matter, including allergens, brought inside on clothes, footwear, pets or by the wind, and that originating indoors from cooking, smoking and the wear and tear of furnishings, are frequently major contributing sources in indoor grit, too as a varied assortment of inorganic and organic chemicals from sources including cleaning products and building materials (Blanchard et al., 2020; Salthammer, 2020). This indoor dust serves as a reservoir for ecology exposure to microbial communities, many of which are benign, some are beneficial, whilst some exhibit pathogenicity. Indeed, the home environment presents an intricate microbial ecosystem inhabited by a typically diverse community of microorganisms, with 500–1,000 different species being reported in house dust (Lax et al., 2014; Barberán et al., 2015; Shan et al., 2019). While many studies describe spatial patterns of bacterial diversity in a specific geographic region or environmental habitat (such as soil, water, or sediment), our knowledge of indoor residential bacterial biodiversity, biogeography and their associated drivers are nonetheless poorly understood.
Local climate and outdoor environmental conditions play a role in the development of the indoor microbiome (Shan et al., 2019). Indoor fungal communities are known to exist strongly influenced by the outdoor environment, with clear geographic patterns reported in the literature (Amend et al., 2010), just the relationship is less clear for indoor microbial communities (Meadow et al., 2014; Barberán et al., 2015). Anthropogenic factors influencing indoor microbial communities include variations in occupancy (both humans and animals), physiological differences in the occupants (east.g. age, gender), lifestyle differences (eastward.grand. diet, living conditions), besides as activity patterns within the home environment (Leung and Lee, 2016). Some studies report Gram-positive phyla such as Firmicutes and Actinobacteria in greater affluence indoors (Täubel et al., 2009; Hanson et al., 2016; Loo et al., 2018), whilst other investigations detect the changed trend with members of the Gram-negative phylum Proteobacteria in greater numbers (Barberán et al., 2015).
Whilst non-occupational exposure to a range of metal(loid)s and organic contaminants in firm dust are a known risk factor for a range of diseases and poor health outcomes (Salthammer, 2020), we know far less virtually the microbial communities associated with our indoor habitation environments, and their interaction/impacts on human health. The majority of research has focussed on selected microbial taxa, investigating affliction manual and pathogenesis within indoor environments, with a focus on impacts on the gastrointestinal (GI) tract and atopy (Haysom and Abrupt, 2003). For instance, Bacteroides spp. (eastward.m. B. fragilis) found in indoor dusts tin can interfere with gut colonization by suppressing anti-microbial allowed responses (Round et al., 2011; Hanson et al., 2016). Notwithstanding, our interactions with the micro-organisms native to the indoor surround may too be beneficial, with contempo inquiry indicating an changed association between the multifariousness of these microorganisms and the prevalence of diverse respiratory conditions of a non-communicable nature such equally wheeze, allergic rhinitis and asthma, too as certain skin conditions like atopic eczema (Birzele et al., 2016; Lee et al., 2018).
The SARS-CoV-2 pandemic has resulted in both tiered and national lockdowns across the world, with an increased risk direction strategy emphasised by Governments' advising regular handwashing, surface sanitation, home-schooling and directives to work from home where possible (Public Health England, 2020). These mitigation strategies have raised concerns that increased exposure to the indoor microbiome, in tandem with reduced exposure to microbial variety in the ambient environment, could exacerbate pre-existing health conditions (Edge et al., 2011; Dannemiller et al., 2014; Juel Holst et al., 2020). Our knowledge, however, of the variety and spatial heterogeneity of bacterial assemblages in our residential environment remains express and we need to better characterise the diverseness, similarities and differences in this microbiome between households to assess the benefits or take a chance posed by our indoor house dust microbiome and to empathize our exposure. Citizen science is a potentially powerful arroyo to both enhance-awareness and to access the residential indoor environment, at scale. Our study is function of the Home Biome projection (www.mapmyenvironment.com) a global report investigating a range of biological and chemical constituents in house grit samples collected by citizens themselves. In this proof-of-concept report we focused on three hypotheses. First, if the indoor residential environment provides a distinctive habitat or biome (the "home biome") we would wait to come across a common cadre house dust microbiome represented in the vacuum dust beyond all homes, regardless of bioclimatic region, building type or occupancy. Second, as global bioclimatic regions influence the animate being and flora of that region we expect to see geospatial dependency, or a region-unique core microbiome typified by bioclimatic zone. Third, citizen collected samples from regular household vacuum cleaners can provide a suitable medium for investigation of the microbial communities nowadays in residential homes. To test these hypotheses, nosotros focussed on the bacterial richness, composition and diversity in citizen nerveless vacuum dust samples from residential homes beyond two countries in ii distinct bioclimatic regions (Uk, Oceanic zone, and Hellenic republic, Mediterranean zone). Our findings have of import implications for the collaboration between citizens and scientists for the development of prove-based direction strategies to modify potential benefits and hazards posed past our indoor house dust microbiome.
Methods
Experimental Design
Several methods have been employed in environmental and public health research for the acquisition of grit samples from residential dwellings. Despite the diversity of approaches, the most commonly used techniques are the utilisation of dry and/or moisture wipes and vacuum cleaners, typically with researcher-led sampling campaigns (e.grand. Rasmussen et al., 2013). In the Home Biome study, nosotros adopted a different approach, that of a citizen-led approach whereby participants were asked to submit either their vacuum bag, or a sub-sample. Written sampling guidance and an online video supported the citizen sampling (details in Supplementary Text S1). Whilst vacuum samples caused as part of big-scale citizen-led campaigns accept the disadvantage of beingness of unknown age and verbal provenance, and collected using a broad array of vacuum devices, enquiry has shown that vacuum cleaner waste is a good surrogate for human exposure to contaminants in indoor dust (Barnes et al., 2013). Indeed vacuum dust samples have been shown to compare well with measurements fabricated past other sampling techniques, likewise equally providing samples that are typically cogitating of an unabridged residential unit of measurement (Colt et al., 2008).
Twenty-8 vacuum dust samples were selected for this investigation (i vacuum sample from each domicile): 20-ane samples from a national entrada within the United kingdom of great britain and northern ireland, representing a regional spread across the UK (including Northern Republic of ireland), and seven samples from Hellenic republic selected to provide a different bioclimatic zone. Both positive and negative reagent controls were included to ensure sterility throughout the processing and sequencing steps, and a randomly selected sample was run in triplicate (DSUK179).
A wide range of anthropogenic factors are likely to touch the indoor microbiome and to capture some of this heterogeneity we developed an online questionnaire which citizens were asked to complete. The questionnaire was split into two sections dealing with 1) the main characteristics and activities of the household occupants, such every bit the number, gender and ages of occupants, pets, the areas of usage of the vacuum cleaner, whether shoes that are worn outside the house were worn indoors, smoking, hobbies, and 2) the residential environment, such as building type and age, main type of flooring roofing, access to outside space and the nature of local surroundings, etc.
Sample Processing and Deoxyribonucleic acid Extraction
On receipt, the samples were logged and stored, unopened, in the dark at room temperature. Sample numberless were opened and the contents sieved in a Class II microbiological safety cabinet using a one-utilise-only 250 μm nylon mesh filter to remove larger particulates and fibrous fabric; the mesh was UV sterilised for >30 min prior to apply. The <250 μm particle size fraction is the fraction normally selected for the chemical analysis of dusts, sediments and soils equally this fraction is more probable to adhere to hands or food produce and exist transferred by ingestion through hand-to-oral cavity contact compared to larger size fractions (Duggan et al., 1986). Sieved particulates were stored in fifty ml sterile falcon tubes until DNA extraction.
High throughput isolation of microbial genomic DNA from each grit sample was performed as per manufacturer'south instructions using a DNeasy 96 PowerSoil Pro Kit (384) (QIAGEN). The DNeasy 96 PowerSoil Pro Kit includes an inhibitor removal stride to eliminate inhibitors commonly plant in soil and ecology samples. Deoxyribonucleic acid samples were stored at −20°C until sequencing.
Sequencing and Data Processing
Taxonomic investigation of leaner was performed via paired-stop sequencing of the V4 hypervariable region of the 16S rRNA gene using the 515-F (GTGCCAGCMGCCGCGGTAA) and 806-R (GGACTACHVGGGTWTCTAAT) primer pair, using a Illumina MiSeq 250 × 2 chemical science approach. Raw sequences were quality checked and trimmed using FastQC/MultiQC and BBDuk (Andrews, 2010; B B Map, 2020). Paired reads were classified using Kraken2 (Wood et al., 2019) and Bracken (Lu et al., 2017) using the SILVA v138 database (Pruesse et al., 2007). Resulting sample report files were converted to BIOM (version ane.0) format (McDonald et al., 2012) using kraken-biom and imported into R (v4.0.3) (R Evolution Core Team, 2020) using the R package Phyloseq (McMurdie and Holmes, 2013). All plots, unless otherwise stated, were produced using GGPlot2 (Wickham, 2009) and organised using Cowplot (Wilke, 2020), GGPubr (Kassambara, 2020) and GGRepel (Slowikowski, 2020). Due to presence in reagent negative control, the genus Escherichia-Shigella was removed from all samples prior to analysis every bit we cannot differentiate its presence as being in the environs sampled or as a result of laboratory exercise and/or contaminated reagents. Afterwards sequence filtering and quality command of the 28 house dust samples, a total of 730,735 sequence reads, with a median/boilerplate of 22,519/25,198 sequence reads per sample (min 8,695, max 45,370) were produced. This corresponded to 600 taxa. The human relationship between number of operational taxonomic units (OTUs) and number of sequences presented is indicative of sufficient sampling depth, increasing sequencing depth farther would provide diminishing returns on newly discovered OTUs. Controls and low read count samples (only 1 sample, DSUK182) were removed, followed past rarefaction (without replacement) to 90% of the minimum sample read count (seven,826 reads per sample) to normalise the library size across the samples. Deoxyribonucleic acid sequencing data consist of discrete counts of sequence reads and the total number of which is the library size (Cameron et al., 2020). Library sizes tin vary greatly between samples and thus the samples were normalised to remove bias and false inferences due to variations in library size. Afterwards rarefaction, 599 unique taxa were obtained beyond all samples and just ane OTU was no longer present in any sample later on random subsampling.
Data Analyses
Due to the inherent complication of the samples, initial abundance profiling was undertaken using relative abundance, calculated and plotted at the phylum level. For phylum level abundance profiling, phyla were included if abundance was greater than 0.5% in at to the lowest degree 25% of all samples, with all other phyla nerveless into an "other" category using the role aggregate_rare of the "Microbiome" R parcel (Lahti and Shetty, 2017). Although an indicator of overall trends, phylum level analysis may not offer sufficient resolution to infer pregnant differences in bacterial communities. As such the phylum level relative affluence profiles were further investigated at family level, including blastoff (i.east. variation of phyla in a single sample) and beta (i.due east. variation of phyla between samples) diversity indices, whilst genus level was used for determinations of similarity per centum analysis (SIMPER).
When the taxa resolution was increased to family unit level, families were included if abundance was greater than two% in as least 50% of the samples, otherwise they were collated as "other". Alpha diversity of the rarefied dataset at family unit level was quantified by using Shannon and Inverse Simpson multifariousness indices, both of which relate OTU richness and evenness and the full number of observed species. Alpha variety metrics were calculated using Phyloseq and statistical significance of observed differences was evaluated using Wilcoxon rank sum exam via the GGPubr parcel office stat_compare_means. Bray-Curtis beta diversity was calculated using vegdist and two-dimension non-metric multidimensional scaling (NMDS) ordination of beta-diversity altitude matrix produced using the function metaMDS every bit office of the "Vegan" R package (Oksanen et al., 2020). Statistical significance of differences in beta-diversity betwixt locations was tested by permutational multivariate assay of variance (PERMANOVA) with the adonis function (Oksanen et al., 2020).
To place both the unique and the shared genera components of the UK and Greek core microbiomes SIMPER analysis was performed at the genus level using the simper function (office of the "Vegan" R package) through the simper_pretty wrapper script (Asteinberger9, 2020). Individual genera identified through this assay were evaluated for location differences based on their relative abundances using a Kruskal-Wallis test via the kruskal. pretty wrapper script (Asteinberger9, 2020). Both the unique and the shared genera components of the cadre microbiomes in the United kingdom and the Greek samples were visualised using the plot_core function of Microbiome (Lahti and Shetty, 2017). To be defined as part of the core microbiome in a location, genera had to exist nowadays at minimum 1% abundance in at least 75% of samples, as previously employed in microbiome analyses (Fahimipour et al., 2018; Rodrigues et al., 2018).
Results and Discussion
Affluence Profiling
At the phylum level the affluence profiles in the bulk of samples from both countries are dominated by Proteobacteria, Actinobacteriota, Firmicutes and Bacteriodota, often accounting for >90% of the relative composition (Supplementary Effigy S1). This correlates with other recent indoor microbiome analyses (Kembel et al., 2014; Gilbert and Stephens, 2018; Nygaard and Charnock, 2018) roofing educational facilities, residential spaces and other common indoor environments, with the key phyla associated with human commensals, and ordinarily found ecology bacteria.
The phylum-level relative abundance profiles were farther investigated at family level (Supplementary Effigy S2). A wide bacterial diversity is evident within each dust sample. Our study focussed on dust samples provided past participants from vacuumed indoor locations inside their own homes. Thus, rather than restricting sampling to specific surfaces, in a specific room, our results reflect the core microbiome across all the vacuumed locations of a participant's habitation with the wide bacterial diversity suggesting the vacuum dust sample does indeed represent a sample from across a range of environments with the home. The peak 8 families identified across the whole dataset include Acetobacteraceae, Chroococcidiopsaceae, Nocardioidaceae, Moraxellaceae, Rhodobacteraceae, Sphingomonadaceae, Staphylococcaceae and Streptococcaceae. The metadata captured by our household questionnaire indicates that a range of unlike abode environments were represented past the grit samples (48% of homes were semi-detached, 37% discrete and 15% units or flats; >seventy% were brick built; 48% were less than 50 years one-time, with 11% older than 100 years; 67% of households had three or more people living in the property, 33% had pets (all UK) and nineteen% had smokers), withal the limited sample size inside each sub-category prevented robust analysis of the influences of these factors on the home microbiome as part of this study (Supplementary Table S1). Whilst characteristics of the domicile, the occupants and their lifestyles, airflow and commutation of textile across the indoor-outdoor interface are all likely to alter the community composition, these viii about abundant families compare with the findings of previous studies across a range of indoor microbiomes and human-associated microbiomes (Barberán et al., 2015; Ding et al., 2020), re-enforcing the link betwixt humans and the bacterial ecology of the indoor microbiome.
Outlier individuals include Prevotellacae (29.46% relative abundance in DSUK013) and Enterobacteriaceae (23.39% relative affluence in DSUK167), both commonly human being associated groups, whilst the relative affluence data for the Greek samples appears to exist skewed by the presence of Rhizobiaceae bookkeeping for up to 50.12% relative affluence in DSGR144, with a group average of 19.xx%. These outliers are specific to certain samples and have likely arisen from specific factors associated with the individual habitation. This warrants farther investigation as nosotros now move to scale upwards this study globally.
Alpha and Beta and Diversity
Family level blastoff diversity distribution of samples, based upon location (UK vs. Greece), was performed and Wilcoxon rank-sum pairwise comparisons identified significant difference between the two countries (Wilcoxon, p = 0.00023 Shannon diversity indices and Wilcoxon, p = 0.00072 Inverse Simpson diversity indices; Supplementary Figure S3). Both the Shannon and Inverse Simpson diversity indices advise well balanced UK communities with regards to abundance and diversity, whilst the Greek samples indicated a lower average diversity (Supplementary Table S2) and may suggest some caste of skewness in the dataset, probable due to a high abundance of Rhizobiaceae_uncultured in the samples.
To investigate the variation in taxonomic abundance profiles between the unlike samples, a 2-dimension NMDS ordination of beta-diversity distance matrix was performed using a Bray-Curtis dissimilarity index (Figure 1). NMDS ordination functions utilize the proximity betwixt objects, which corresponds to their similarity, to differentiate between samples. Statistically meaning differences were observed (PERMANOVA F-value: iii.5807, R-squared: 0.11709, p-value 0.001; NMDS Stress = 0.144). Figure 1 clearly differentiates the samples and clusters according to the location metadata grouping of U.k. vs. Hellenic republic.
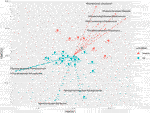
FIGURE 1. Beta Diversity NMDS bi-plot using Bray distance; vector lines testify FDR-adjusted statistically significant taxa that contribute to ordination. Samples are grouped by location.
Characterization of a Mutual and a Region-Specific Core Indoor Microbiome
With appreciable trends apparent in the beta diversity identified using PERMANOVA, the outcome of location was further explored at genus level using FDR-adjusted statistical methods. The contribution of individual OTUs to the genus level Bray-Curtis beta diversity was performed using SIMPER analysis to categorise most arable OTUs using a pairwise comparison of the UK vs. Greece dataset at the genus level. SIMPER analysis cumulatively accounts 100% of the variation between the comparison of the United kingdom vs. Greece, every bit identified using the Bray-Curtis measures. Tabular array one lists 20 taxa responsible for >one% variation between the ii countries, of which 9 taxa were identified to be statistically significant by comparing of OTU relative abundance by location metadata using Kruskal-Wallis rank sum testing with additional FDR-adjustments. To evaluate the genera contribution to ordination, the Vegan function envfit (999 permutations) was used to add together species-loading vectors to the NMDS biplot (Figure 1), with vectors shown for those genera determined to be FDR-adjusted statistically significant from Tabular array 1.

Table one. SIMPER assay of significant taxa driving diversity at genus level. Significance testing performed using Kruskal-Wallis one-way analysis of variance bookkeeping for false discovery rates. Greyed out rows fail to run across the statistical significance cutting off value of p < 0.05. H in parentheses denotes a ordinarily human associated Genus.
These FDR adjusted statistically significant taxa are presented in Effigy 2, highlighting the relative abundance profiles between location groups (UK vs. Hellenic republic). The taxon labelled Rhizobiaceae_uncultured by the SILVA database appears to be the single most abundant bacteria responsible for diversity between samples contributing 15.41% of the dissimilarity, with other private taxa responsible for not more than iv.50% contrast. Notably, this taxon is not described in the family level relative abundance profiling described earlier (Supplementary Figure S2) but is considered during the examination of notable outliers. As affluence values are skewed from the loftier prevalence of this taxa in only a select number of Greek samples, in combination with low prevalence within UK samples, Rhizobiaceae_uncultured is not considered a fellow member of the overall most abundant taxa.

FIGURE 2. SIMPER analysis violin plots showing the statistically significant FDR adapted p value taxa from Table one. Contributing SIMPER and Kruskal Wallis (KW) values are displayed for each taxon, and by country.
An investigation into the cadre taxa was performed using a robust taxa prevalence threshold of 75% at a i% detection (Effigy 3), a threshold previously recommended (Fahimipour et al., 2018; Rodrigues et al., 2018), with the aim to place both a set of genera common to all of the indoor dust samples (a shared common cadre microbiome), and also a set of genera that are unique to either all of the UK samples, or to all of the Greek samples (a unique core microbiome). To preserve the abundance values of the cadre microbiome between locations, the shared cadre microbiome was plotted independently in improver to location specific unique taxa (Figure 3).
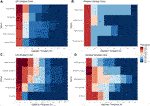
FIGURE 3. A core microbiome assay roofing unique and shared taxa for each location based upon a robust 75% prevalence threshold with at least 1% detection threshold. (A) UK unique core microbiome (B) Hellenic republic unique core microbiome, (C) UK shared core microbiome, (D) Greece shared cadre microbiome.
The shared core microbiome (i.e. observed throughout all of the indoor dust samples) included Massalia, Acinetobacter, Staphylococcus, Rubellimicrobium, and Sphingomonas (Figures 3C,D). Interestingly, two of these genera (Sphingomonas and Massilia) also contributed to the observed significant variation based upon geospatial location using FDR-adjusted Kruskal Wallis rank sum testing (Effigy 1; p Sphingomonas = 0.0312 and p Massilia = 0.0198). This indicates that while both are core genera found in firm grit, Sphingomonas have a higher contribution to percentage taxonomic abundance in UK dust samples, conversely to Massilia which has a higher percentage affluence in samples from Greece (Effigy 2). Sphingomonas (Family unit Spirosomaceae) has been found in both aqueous and terrestrial habitats, including institute root systems, clinical specimens, and other mutual contexts. While Massilia, a member the family unit Oxalobacteraceae, is typically found in a diverse range of environmental habitats and is normally reported as an air, h2o, soil, and plant-associated bacterium (Chaudhary and Kim, 2017; Frediansyah et al., 2020).
The other identified members of this shared core microbiome accept been frequently reported in non-residential indoor microbiomes. Acinetobacter are a strictly aerobic genera of microbes, known to reside in a broad variety of environments including soil and wastewater, cause contamination of food products, and are a known commensal bacterium residing on the skin and mucosal membranes. Previously considered saprophytic with lilliputian clinical significance, simply with the emergence of antimicrobials in both agronomical and clinical settings, Acinetobacter spp. have become of increasing concern due farthermost drug resistance, thereby impeding therapeutic treatments (Doughari et al., 2011). Importantly, the bacteria tin besides colonise the peel and respiratory tract without causing an infection, with subsequent infections occurring if the hosts primary line of defence force is compromised. Staphylcoccus is a genus of Gram-positive bacteria in the family Staphylococcaceae from the order Bacillales. There are currently at least twoscore species inside the genus. Many are unable to cause disease, but one of the nigh well described species is Staphylococcus aureus, a commensal of the human microbiota that is normally labelled every bit an opportunistic pathogen capable of causing a range of skin and respiratory infections. It is estimated that between 20–30% of the homo population are carriers of Due south. aureus (Tong et al., 2015). Rubellimicrobium appears to be an nether described bacterium normally isolated from environments including soil and air (Jiang et al., 2019).
Microbes assigned every bit "unique core" taxa from Hellenic republic include Paracoccus, Blastococcus, Deinococcus and Pseudomonas (Effigy 3B); Paracoccus, Blastococcus and Deinococcus were identified to be statistically meaning contributors to the observed ordination diversity (Figure i; FDR-adjusted Kruskal Wallis: p Blastococcus = 0.013, p Deinococcus = 0.017, p Paracoccus = 0.031), indicating their contribution to the variety. Significantly, all three taxa are associated with arid landscapes, such every bit occur in the Mediterranean climate of Greece, in contrast to the Oceanic/Maritime conditions of the UK (Baker et al., 1998; Lasek et al., 2018; Makarova et al., 2001; Urzì et al., 2001; Zhu et al., 2013). The UK unique core taxa (Figure 3A) included Nocardiodes, Spirosoma, Streptococcus, Corynebacterium and Hymenobacter, of which Nocardiodes and Spirosoma were identified to be statistically significant contributors to the observed ordination variety (Figure 1; FDR-adjusted Kruskal Wallis: p Spirosoma = 0.002 p Nocardioides = 0.013), once more indicating their contribution to the diversity. This unique UK core consists of homo commensals and environmental bacteria typically associated with soils (Yoon and Park, 2006; Jeon et al., 2013; Kim et al., 2016; Leal et al., 2016). The depression prevalence of the UK unique cadre microbes in the samples from Hellenic republic and the low prevalence of the Greece unique core microbes in the samples from the U.k. further demonstrates the difference (Supplementary Effigy S4). Exploration into the microbial diversity identified that all samples with sufficient read depth showed splendid richness and diverseness, with a few notable exceptions of taxa dominating particular samples, including an undefined Rhizobiaceae (49.96% in sample DSGR144; 41.74% in sample DSGR146) also every bit Prevotellacae (29.46% in sample DSUK013).
Written report Limitations
Whilst the written report is express by its geographical extent (only ii bioclimatic regions), and the number of participating households (20-eight), insights gained from this feasibility written report are informing our scaling-upwardly activities equally a much larger study is required to determine the contribution of building type, occupancy level, building fabric, etc. to the overall indoor microbiome. Although we fabricated efforts to use articulate, unambiguous linguistic communication to support the sampling and questionnaire survey, missing data on participant vacuum sampling date and variations in interpretation of some of the survey questions required a re-design of selected questions and procedures. For example, our endeavor to capture both the "age" of the vacuum sample (i.due east. time period of vacuuming represented past the sample) and the primary home construction material was inconsistently interpreted/reported (e.g. some householders reported multiple home fabric, e.g. UK166 and 167, Supplementary Tabular array S1). Such challenges are a well-recognised aspect of citizen science, but the opportunities afforded by such collaborations are increasingly beingness realised and best-selling (Phillips et al., 2019).
Conclusion
Our study showed that citizen collected samples from regular household vacuum cleaners tin provide a suitable medium for investigation of the microbial communities nowadays in residential homes. Diverse household samples, regardless of bioclimatic region, edifice type or occupancy, were shown to share a mutual core microbiome consisting of Sphingomonas, Rubellimicrobium, Staphylococcus, Acinetobacter and Massalia. Indeed, despite clear differences existence reported in the homo microbiome (which contributes directly to the house dust samples), a core "dwelling house microbiome" could still be detected.
Several of the bacterial genera identified as common in the home microbiome are environmentally ubiquitous (east.grand., Acinetobacter, Staphylococcus, Corynebacterium) and whilst pathogenicity is species/strain-specific, and also dependent on the immune-status of the host, some similar Acinetobacter spp. are increasingly of interest due to farthermost drug resistance (Doughari et al., 2011). Occurrence as part of the core dwelling house microbiome is of relevance and highlights cardinal genera for targeted farther work to establish species-level identification.
Whilst several of the prominent taxa identified in the microbial communities of our household dusts were dominated past both human and non-human commensal bacteria, we likewise identified the presence of a region-specific core microbiome. This geospatial dependency may reflect differences in our indoor bacterial communities due to prevailing bioclimatic or country-specific factors (e.thou. Paracoccus, Blastococcus, Deinococcus and Pseudomonas in the Greek samples, and Nocardiodes, Spirosoma, Streptococcus, Corynebacterium and Hymenobacter in the United kingdom of great britain and northern ireland unique core). We at present need to scale up this study to robustly test these hypotheses. To further understand the differences between homes, future studies should also consider including analysis of the microbiomes of the occupants (both human and animal), in parallel with key characteristics of the local surround such as the composition of the local soils, given they are key sources of input to house dust.
Data Availability Statement
The sequencing data that support the findings of this study are openly bachelor (every bit raw fastq files: Target_gene 16S and Target_subfragment V4) from the European Nucleotide Archive via the study accretion PRJEB46920 (individual sample accretion numbers ERX6130460 to ERX6130493). URL: https://www.ebi.air-conditioning.uk/ena/browser/view/PRJEB46920.
Ethics
This report was part of the Domicile Biome Projection, ethics approval number: 19135, Department of Geography and Ecology Scientific discipline Ethics Committee, Northumbria University, UK.
Author Contributions
JE, DP, AH, AN, JW, PR, SP-V, LB, AA, and PM conceptualised the Home Biome project, input into the experimental blueprint, and input to the manuscript. JT initiated the analyses, designed and performed the experiments and prepared the initial draft manuscript. MC adult the bioinformatic and statistical analysis pipeline, MB brash on bioinformatics analysis. I managed the in-house sequencing. AA provided the Greek samples. KJ was involved in experimental design and literature review. All authors contributed to and approved the final manuscript.
Funding
JE acknowledges funding from the Natural Environment Research Quango (Enquiry Grant NE/T004401/1). MB and MC admit enquiry funding from Research England'south Expanding Excellence in England (E3) Fund as office of the Hub for Biotechnology in the Built Environment. KJ acknowledges funding for her PhD research programme from Northumbria Academy. Open admission publication fees were provide by Northumbria University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of involvement.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily stand for those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this commodity, or claim that may exist made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Cloth
The Supplementary Fabric for this article can be found online at: https://www.frontiersin.org/articles/x.3389/fenvs.2021.754657/full#supplementary-cloth
References
Better, A. South., Seifert, Chiliad. A., Samson, R., and Bruns, T. D. (2010). Indoor Fungal Composition Is Geographically Patterned and More Diverse in Temperate Zones Than in the Tropics. Proc. Natl. Acad. Sci. 107, 13748–13753. doi:x.1073/pnas.1000454107
PubMed Abstract | CrossRef Full Text | Google Scholar
Baker, Due south. C., Ferguson, South. J., Ludwig, B., Page, M. D., Richter, O.-Yard. H., and van Spanning, R. J. M. (1998). Molecular Genetics of the Genus Paracoccus : Metabolically Versatile Bacteria with Bioenergetic Flexibility. Microbiol. Mol. Biol. Rev. 62 (iv), 1046–1078. doi:10.1128/mmbr.62.four.1046-1078.1998
PubMed Abstract | CrossRef Full Text | Google Scholar
Barberán, A., Dunn, R. R., Reich, B. J., Pacifici, K., Laber, E. B., Menninger, H. L., et al. (2015). The Environmental of Microscopic Life in Household Dust. Proc. R. Soc. B. 282, 20151139. doi:ten.1098/rspb.2015.1139
CrossRef Total Text | Google Scholar
Barnes, C., Portnoy, J. M., Ciaccio, C. Eastward., and Pacheco, F. (2013). A Comparison of Subject Room Dust with home Vacuum Grit for Evaluation of Dust-Borne Aeroallergens. Ann. Allergy Asthma Immunol. 110 (v), 375–379. doi:10.1016/j.anai.2013.02.010
PubMed Abstract | CrossRef Total Text | Google Scholar
Birzele, Fifty. T., Depner, One thousand., Ege, M. J., Engel, M., Kublik, Due south., Bernau, C., et al. (2016). Environmental and Mucosal Microbiota and Their Role in Babyhood Asthma. Allergy 72 (i), 109–119. doi:10.1111/all.13002
PubMed Abstract | CrossRef Full Text | Google Scholar
Blanchard, P., Babichuk, N., and Sarkar, A. (2020). Evaluating the Employ of Synchrotron 10-ray Spectroscopy in Investigating Brominated Flame Retardants in Indoor Grit. Environ. Sci. Pollut. Res. 27 (33), 42168–42174. doi:10.1007/s11356-020-10623-4
CrossRef Full Text | Google Scholar
Cameron, Due east. South., Schmidt, P. J., Tremblay, B. J., Emelko, M. B., and Müller, K. M. (2020). To Rarefy or Non to Rarefy: Enhancing Microbial Community Analysis through Next-Generation Sequencing. bioRxiv. doi:10.1101/2020.09.09.290049
CrossRef Full Text | Google Scholar
Chaudhary, D. K., and Kim, J. (2017). Massilia Agri Sp. nov., Isolated from Reclaimed Grassland Soil. Int. J. Syst. Evol. Microbiol. 67 (8), 2696–2703. doi:10.1099/ijsem.0.002002
CrossRef Total Text | Google Scholar
Colt, J. Due south., Gunier, R. B., Metayer, C., Nishioka, One thousand. G., Bell, East. M., Reynolds, P., et al. (2008). Household Vacuum Cleaners vs. The High-Book Surface Sampler for Drove of Rug Dust Samples in Epidemiologic Studies of Children. Environ. Wellness 7, half dozen. doi:x.1186/1476-069X-vii-6
PubMed Abstract | CrossRef Total Text | Google Scholar
Dannemiller, K. C., Mendell, 1000. J., Macher, J. M., Kumagai, K., Bradman, A., The netherlands, N., et al. (2014). Next-generation Deoxyribonucleic acid Sequencing Reveals that Low Fungal Diversity in House Dust Is Associated with Childhood Asthma Development. Indoor Air 24 (3), 236–247. doi:x.1111/ina.12072
PubMed Abstruse | CrossRef Total Text | Google Scholar
Ding, 50.-J., Zhou, X.-Y., and Zhu, Y.-G. (2020). Microbiome and Antibiotic Resistome in Household Dust from Beijing, China. Environ. Int. 139, 105702. doi:x.1016/j.envint.2020.105702
PubMed Abstruse | CrossRef Full Text | Google Scholar
Doughari, H. J., Ndakidemi, P. A., Human, I. S., and Benade, S. (2011). The Environmental, Biological science and Pathogenesis of Acinetobacter spp.: an Overview. Microb. Environ. 26 (2), 101–112. doi:10.1264/jsme2.me10179
PubMed Abstract | CrossRef Full Text | Google Scholar
Duggan, M. J., Inskip, Chiliad. J., Rundle, S. A., and Moorcroft, J. Southward. (1986). Pb in Playground Dust and on the Easily of Schoolchildren. Sci. Total Environ. 44 (i), 65–79. doi:10.1016/0048-9697(85)90051-8
CrossRef Full Text | Google Scholar
Ege, M. J., Mayer, M., Normand, A. C., Genuneit, J., Cookson, West. O., Braun-Fahrländer, C., et al. (2011). Exposure to Environmental Microorganisms and Childhood Asthma. North. Engl. J. Med. 364 (8), 701–709. doi:10.1056/NEJMoa1007302
CrossRef Full Text | Google Scholar
Fahimipour, A. One thousand., Ben Mamaar, S., McFarland, A. Thou., Blaustein, R. A., Chen, J., Glawe, A. J., et al. (2018). Antimicrobial Chemicals Associate with Microbial Function and Antibiotic Resistance Indoors. mSystems iii (half-dozen), e00200–18. doi:10.1128/mSystems.00200-eighteen
PubMed Abstract | CrossRef Full Text | Google Scholar
Frediansyah, A., Straetener, J., Brötz-Oesterhelt, H., and Gross, H. (2020). Massiliamide, a Cyclic Tetrapeptide with Potent Tyrosinase Inhibitory Properties from the Gram-Negative Bacterium Massilia Albidiflava DSM 17472T. J. Antibiot. 74 (4), 269–272. doi:10.1038/s41429-020-00394-y
CrossRef Full Text | Google Scholar
Hanson, B., Zhou, Y., Bautista, E. J., Urch, B., Speck, Yard., Silverman, F., et al. (2016). Label of the Bacterial and Fungal Microbiome in Indoor Dust and Outdoor Air Samples: a Airplane pilot Study. Environ. Sci. Procedure. Impacts 18 (6), 713–724. doi:10.1039/c5em00639b
PubMed Abstract | CrossRef Full Text | Google Scholar
Haysom, I. W., and Sharp, K. (2003). The Survival and Recovery of Bacteria in Vacuum Cleaner Dust. J. R. Soc. Promot. Health 123 (ane), 39–45. doi:10.1177/146642400312300114
CrossRef Full Text | Google Scholar
Jeon, Y.-S., Chun, J., and Kim, B.-S. (2013). Identification of Household Bacterial Community and Analysis of Species Shared with Human Microbiome. Curr. Microbiol. 67 (v), 557–563. doi:ten.1007/s00284-013-0401-y
PubMed Abstract | CrossRef Total Text | Google Scholar
Jiang, L.-Q., Zhang, K., Li, Thou.-D., Wang, Ten.-Y., Shi, Due south.-B., Li, Q.-Y., et al. (2019). Rubellimicrobium Rubrum Sp. nov., a Novel Bright Reddish Bacterium Isolated from a Lichen Sample. Antonie van Leeuwenhoek 112 (12), 1739–1745. doi:ten.1007/s10482-019-01304-5
PubMed Abstract | CrossRef Full Text | Google Scholar
Juel Holst, G., Pørneki, A., Lindgreen, J., Thuesen, B., Bønløkke, J., Hyvärinen, A., et al. (2020). Household Dampness and Microbial Exposure Related to Allergy and Respiratory Health in Danish Adults. Eur. Clin. Respir. J. 7 (1), 1706235. doi:ten.1080/20018525.2019.1706235
PubMed Abstruse | CrossRef Full Text | Google Scholar
Kembel, S. W., Meadow, J. F., O'Connor, T. K., Mhuireach, G., Northcutt, D., Kline, J., et al. (2014). Architectural Blueprint Drives the Biogeography of Indoor Bacterial Communities. PLOS ONE 9 (1), e87093. doi:10.1371/periodical.pone.0087093
PubMed Abstract | CrossRef Full Text | Google Scholar
Kim, S.-J., Ahn, J.-H., Weon, H.-Y., Hong, S.-B., Seok, Due south.-J., Kim, J.-S., et al. (2016). Spirosoma Aerophilum Sp. nov., Isolated from an Air Sample. Int. J. Syst. Evol. Microbiol. 66 (6), 2342–2346. doi:x.1099/ijsem.0.001037
CrossRef Full Text | Google Scholar
Lasek, R., Szuplewska, M., Mitura, M., Decewicz, P., Chmielowska, C., Pawłot, A., et al. (2018). Genome Construction of the Opportunistic Pathogen Paracoccus Yeei (Alphaproteobacteria) and Identification of Putative Virulence Factors. Front. Microbiol. 9, 2553. doi:10.3389/fmicb.2018.02553
PubMed Abstract | CrossRef Full Text | Google Scholar
Lax, S., Smith, D. P., Hampton-Marcell, J., Owens, South. Grand., Handley, K. 1000., Scott, Due north. Thousand., et al. (2014). Longitudinal Analysis of Microbial Interaction between Humans and the Indoor Surround. Science 345 (6200), 1048–1052. doi:10.1126/scientific discipline.1254529
PubMed Abstract | CrossRef Full Text | Google Scholar
Leal, S. Yard., Jones, Grand., and Gilligan, P. H. (2016). Clinical Significance of Commensal Gram-Positive Rods Routinely Isolated from Patient Samples. J. Clin. Microbiol. 54 (12), 2928–2936. doi:x.1128/jcm.01393-16
PubMed Abstract | CrossRef Full Text | Google Scholar
Lee, Thousand. K., Carnes, M. U., Butz, N., Azcarate-Peril, Chiliad. A., Richards, M., Umbach, D. Thousand., et al. (2018). Exposures Related to House Grit Microbiota in a U.S. Farming Population. Environ. Health Perspect. 126 (6), 067001. doi:10.1289/ehp3145
PubMed Abstruse | CrossRef Total Text | Google Scholar
Leung, M. H. Y., and Lee, P. K. H. (2016). The Roles of the Outdoors and Occupants in Contributing to a Potential Pan-Microbiome of the Built Environment: a Review. Microbiome 4 (ane), 21. doi:10.1186/s40168-016-0165-2
PubMed Abstract | CrossRef Full Text | Google Scholar
Loo, Eastward. 10. Fifty., Chew, L. J. G., Zulkifli, A. B., Ta, L. D. H., Kuo, I.-C., Goh, A., et al. (2018). Comparison of Microbiota and Allergen Profile in House Grit from Homes of Allergic and Non-allergic Subjects- Results from the GUSTO Study. World Allergy Organ. J. xi, 37. doi:x.1186/s40413-018-0212-five
PubMed Abstract | CrossRef Full Text | Google Scholar
Lu, J., Breitwieser, F. P., Thielen, P., and Salzberg, S. Fifty. (2017). Bracken: Estimating Species Abundance in Metagenomics Information. Peer J. Comput. Sci. 3, e104. doi:10.7717/peerj-cs.104
CrossRef Full Text | Google Scholar
Makarova, G. S., Aravind, L., Wolf, Y. I., Tatusov, R. L., Minton, Grand. West., Koonin, Due east. 5., et al. (2001). Genome of the Extremely Radiation-Resistant Bacterium Deinococcus Radiodurans Viewed from the Perspective of Comparative Genomics. Microbiol. Mol. Biol. Rev. 65 (1), 44–79. doi:10.1128/mmbr.65.1.44-79.2001
PubMed Abstract | CrossRef Full Text | Google Scholar
McDonald, D., Clemente, J. C., Kuczynski, J., Rideout, J. R., Stombaugh, J., Wendel, D., et al. (2012). The Biological Observation Matrix (BIOM) Format or: How I Learned to Stop Worrying and Love the Ome-Ome. GigaSci ane (1), vii. doi:x.1186/2047-217X-1-7
PubMed Abstruse | CrossRef Full Text | Google Scholar
McMurdie, P. J., and Holmes, South. (2013). Phyloseq: an R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS I 8 (4), e61217. doi:10.1371/journal.pone.0061217
PubMed Abstract | CrossRef Total Text | Google Scholar
Meadow, J. F., Altrichter, A. Due east., Kembel, S. W., Kline, J., Mhuireach, G., Moriyama, M., et al. (2014). Indoor Airborne Bacterial Communities Are Influenced by Ventilation, Occupancy, and Outdoor Air Source. Indoor Air 24, 41–48. doi:x.1111/ina.12047
PubMed Abstruse | CrossRef Total Text | Google Scholar
Morawska, L., and Salthammer, T. (2003). Indoor Surround: Airborne Particles and Settled Grit. Weinheim: Wiley-VCH, 467. ISBN: 978-3-527-30525-4.
Google Scholar
Nygaard, A. B., and Charnock, C. (2018). Longitudinal Development of the Dust Microbiome in a Newly Opened Norwegian Kindergarten. Microbiome 6 (1), 159. doi:x.1186/s40168-018-0553-x
PubMed Abstruse | CrossRef Full Text | Google Scholar
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2020). Vegan Community Environmental Package Version 2. Bachelor at: https://cran.r-projection.org, https://github.com/vegandevs/vegan (Accessed Nov 5–7)
Google Scholar
Phillips, T. B., Ballard, H. L., Lewenstein, B. V., and Bonney, R. (2019). Appointment in Science through Citizen Science: Moving across Data Collection. Sci. Ed. 103, 665–690. doi:x.1002/sce.21501
CrossRef Full Text | Google Scholar
Pruesse, E., Quast, C., Knittel, 1000., Fuchs, B. Thou., Ludwig, Westward., Peplies, J., et al. (2007). SILVA: a Comprehensive Online Resource for Quality Checked and Aligned Ribosomal RNA Sequence Information Compatible with ARB. Nucleic Acids Res. 35 (21), 7188–7196. doi:10.1093/nar/gkm864
PubMed Abstract | CrossRef Full Text | Google Scholar
R Development Core Team (2020). R: A Language and Surround for Statistical Computing. Vienna, Republic of austria: R Foundation for Statistical Computing.
Google Scholar
Rasmussen, P. Due east., Levesque, C., Chénier, One thousand., Gardner, H. D., Jones-Otazo, H., and Petrovic, S. (2013). Canadian House Dust Written report: Population-Based Concentrations, Loads and Loading Rates of Arsenic, Cadmium, Chromium, Copper, Nickel, atomic number 82, and Zinc inside Urban Homes. Sci. Total Environ. 443, 520–529. doi:10.1016/j.scitotenv.2012.11.003
PubMed Abstruse | CrossRef Full Text | Google Scholar
Circular, J. L., Lee, S. M., Li, J., Tran, One thousand., Jabri, B., Chatila, T. A., et al. (2011). The Cost-like Receptor 2 Pathway Establishes Colonization by a Commensal of the Human Microbiota. Science 332, 974–977. doi:10.1126/science.1206095
PubMed Abstract | CrossRef Full Text | Google Scholar
Shan, Y., Wu, Westward., Fan, Due west., Haahtela, T., and Zhang, G. (2019). House Dust Microbiome and Human Health Risks. Int. Microbiol. 22 (3), 297–304. doi:10.1007/s10123-019-00057-v
PubMed Abstract | CrossRef Total Text | Google Scholar
Täubel, One thousand., Rintala, H., Pitkäranta, M., Paulin, Fifty., Laitinen, South., Pekkanen, J., et al. (2009). The Occupant as a Source of House Dust Leaner. J. Allergy Clin. Immunol. 124 (4), 834–840. doi:10.1016/j.jaci.2009.07.045
CrossRef Full Text | Google Scholar
Tong, Southward. Y. C., Davis, J. Southward., Eichenberger, Eastward., The netherlands, T. Fifty., and Fowler, 5. G. (2015). Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 28 (3), 603–661. doi:10.1128/cmr.00134-14
PubMed Abstruse | CrossRef Full Text | Google Scholar
Urzì, C., Brusetti, Fifty., Salamone, P., Sorlini, C., Stackebrandt, E., and Daffonchio, D. (2001). Biodiversity of Geodermatophilaceae Isolated from Contradistinct Stones and Monuments in the Mediterranean bowl. Environ. Microbiol. 3 (7), 471–479. doi:10.1046/j.1462-2920.2001.00217.x
PubMed Abstract | CrossRef Full Text | Google Scholar
Wickham, H. (2009). ggplot2: Elegant Graphics for Data Analysis. New York: Springer. ISBN 978-0-387-98141-3.
Google Scholar
Yoon, J-H., and Park, Y-H. (2006). "The Genus Nocardioides. P. 1099-113," in Volume iii: The Prokaryotes: Archaea Bacteria: Firmicutes, Actinomycetes. Editors Grand. Dworkin, S. Falkow, Eastward. Rosenberg, K-H. Schleifer, and Eastward. Stackebrandt (New York, NYNew York: Springer). ISBN 978-0-387-30743-5.
Google Scholar
Zhu, W. Y., Zhang, J. Fifty., Qin, Y. L., Xiong, Z. J., Zhang, D. F., Klenk, H. P., et al. (2013). Blastococcus Endophyticus Sp. nov., an Actinobacterium Isolated from Camptotheca Acuminata. Int. J. Syst. Evol. Microbiol. 63 (9), 3269–3273. doi:10.1099/ijs.0.049239-0
PubMed Abstruse | CrossRef Full Text | Google Scholar
Source: https://www.frontiersin.org/articles/10.3389/fenvs.2021.754657/full
0 Response to "Gilbert and Stephens Microbiology of the Built Environment Nature Reviews Microbiology 2018"
Post a Comment